Random Search
This example is using NetLogo Flocking model (Wilensky, 1998) to demonstrate random search for optimal parameter values.
Like in the LHS example the Latin hypercube sampling method is used but instead of categorical criteria the simulations are evaluated with single best-fit criterion defined in Best-fit example.
library(tgp)
experiment <- nl_experiment(
model_file = "models/Sample Models/Biology/Flocking.nlogo",
setup_commands = c("setup", "repeat 100 [go]"),
iterations = 5,
param_values = nl_param_lhs(
n = 100,
world_size = 50,
population = 80,
vision = 6,
min_separation = c(0, 4),
max_align_turn = c(0, 20)
),
mapping = c(
min_separation = "minimum-separation",
max_align_turn = "max-align-turn"),
step_measures = measures(
converged = "1 -
(standard-deviation [dx] of turtles +
standard-deviation [dy] of turtles) / 2",
mean_crowding =
"mean [count flockmates + 1] of turtles"
),
eval_criteria = criteria( # aggregate over time
c_converged = mean(step$converged),
c_mcrowding = mean(step$mean_crowding)
),
repetitions = 10, # repeat simulations 10 times
random_seed = 1:10,
eval_aggregate_fun = mean # aggregate over repetitions
) result <- nl_run(experiment, parallel = TRUE)We are using the same best-fit criterion function as in previous example:
\[ value = \sqrt{(crowding - 8)^2 + 100 \times (converged - 1)^2} \]
dat <- nl_get_criteria_result(
result,
eval_value = sqrt((c_mcrowding - 8)^2 + 400*(c_converged - 1)^2)
)res_value <- min(dat$eval_value)
min_eval_id <- which(dat$eval_value == res_value)
res_params <- dat[min_eval_id, c("min_separation", "max_align_turn")]
round(res_value, 1)
#> [1] 4.1
round(res_params,1)
#> min_separation max_align_turn
#> 16 2.1 18.8dat$eval_id = 1:nrow(result$criteria)
library(ggplot2)
ggplot(dat, aes(x = min_separation, y = max_align_turn, color = eval_value)) +
geom_point(alpha = 0.3, size = 3) +
geom_point(
data = subset(dat, eval_id == min_eval_id), size = 10, shape = 1) +
theme_minimal() +
theme(legend.position = "none")dat$min_eval <-
sapply(1:nrow(dat), function(i) min(dat$eval_value[dat$eval_id <= i]) )
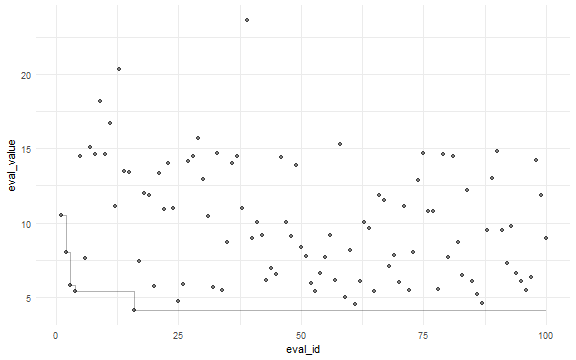
ggplot(dat, aes(x = eval_id, y = eval_value)) +
geom_point(alpha = 0.5) +
geom_step(aes(x = eval_id, y = min_eval), alpha = 0.3) +
theme_minimal()
See also
L-BFGS-B Optimization shows how to use optimization functions from other R packages.
Nelder-Mead Optimization shows how to use optimization functions from other R packages.
Simulated Annealing demonstrates optimization with simulated annealing.
Genetic Algorithm demonstrates optimization with genetic algorithm.
Categorical Criteria and Hyper Latinc Cube Sampling examples demontrate how to explore parameter space with categorical criteria and sampling methods.